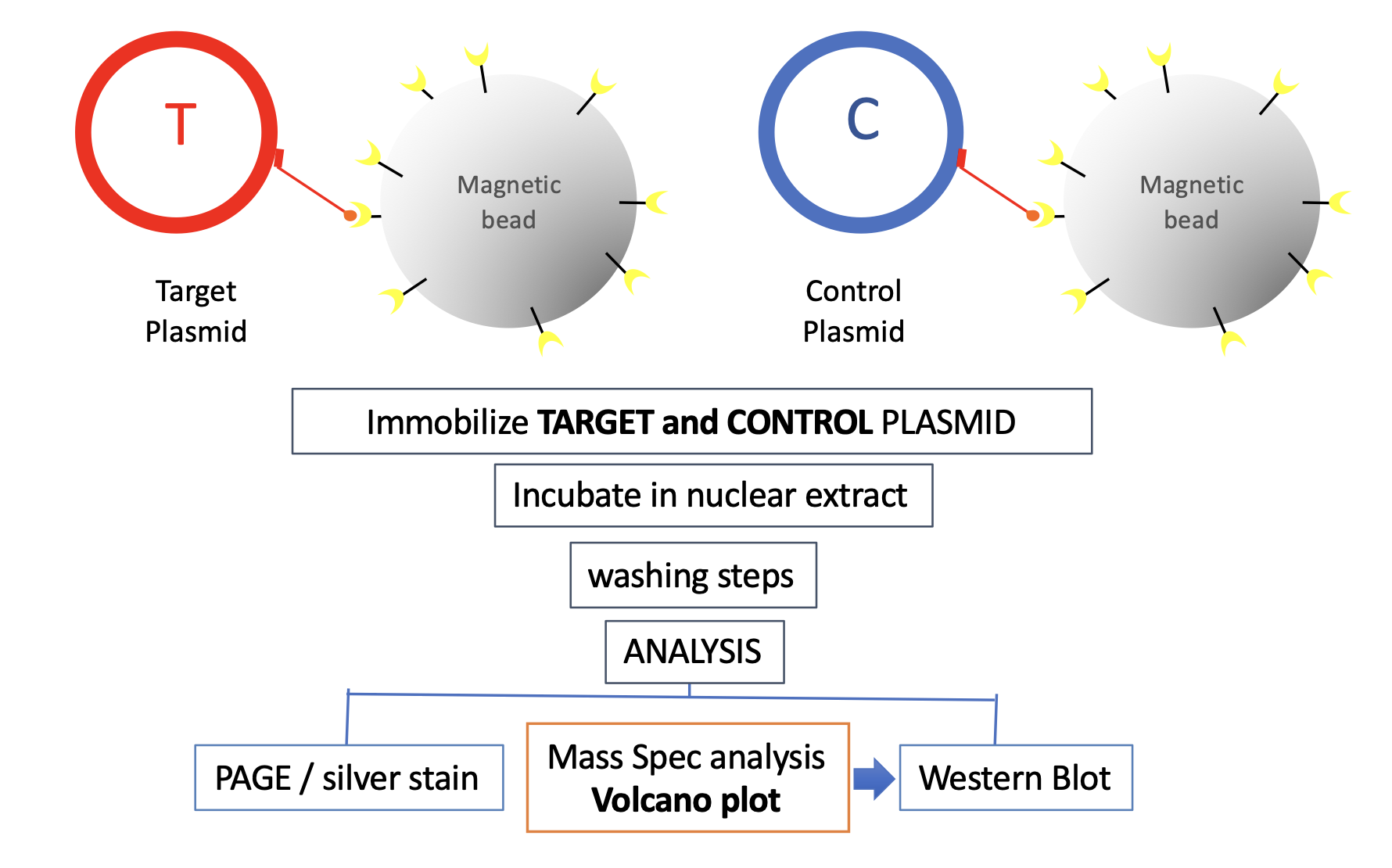
How does IDAP work? As illustrated below, IDAP involves plasmids, derived from a proprietary vector backbone. A Target plasmid that contains a DNA element-of-interest to which one wishes to discover the specific binding proteins and the Control plasmid, devoid of that element, are separately immobilized on streptavidin coated magnetic beads via a triple helix-forming probe. The plasmid-bound beads are incubated in an extract of choice, under defined conditions of time and temperature.
Following incubation, both Target and Control plasmids are recovered on a magnet, submitted to a proprietary washing procedure to remove non-specific proteins. Subsequently, protein assembled on DNA are analyzed by Mass Spectrometry or Western Blotting. Identification of proteins specifically assembled on the DNA element-of-interest is implemented by comparing proteins bound to Target and Control plasmid. The IDAP approach is versatile and robust, however it is a screen that needs further validation of the discovered hits by biochemical and/or genetic experiments.